Abstract
BACKGROUND AND PURPOSE: Privacy concerns, such as identifiable facial features within brain scans, have hindered the availability of pediatric neuroimaging data sets for research. Consequently, pediatric neuroscience research lags adult counterparts, particularly in rare disease and under-represented populations. The removal of face regions (image defacing) can mitigate this; however, existing defacing tools often fail with pediatric cases and diverse image types, leaving a critical gap in data accessibility. Given recent National Institutes of Health data sharing mandates, novel solutions are a critical need.
MATERIALS AND METHODS: To develop an artificial intelligence (AI)-powered tool for automatic defacing of pediatric brain MRIs, deep learning methodologies (nnU-Net) were used by using a large, diverse multi-institutional data set of clinical radiology images. This included multiparametric MRIs (T1-weighted [T1W], T1W-contrast-enhanced, T2-weighted [T2W], T2W-FLAIR) with 976 total images from 208 patients with brain tumor (Children’s Brain Tumor Network, CBTN) and 36 clinical control patients (Scans with Limited Imaging Pathology, SLIP) ranging in age from 7 days to 21 years old.
RESULTS: Face and ear removal accuracy for withheld testing data were the primary measure of model performance. Potential influences of defacing on downstream research usage were evaluated with standard image processing and AI-based pipelines. Group-level statistical trends were compared between original (nondefaced) and defaced images. Across image types, the model had high accuracy for removing face regions (mean accuracy, 98%; n=98 subjects/392 images), with lower performance for removal of ears (73%). Analysis of global and regional brain measures (SLIP cohort) showed minimal differences between original and defaced outputs (mean rS = 0.93, all P < .0001). AI-generated whole brain and tumor volumes (CBTN cohort) and temporalis muscle metrics (volume, cross-sectional area, centile scores; SLIP cohort) were not significantly affected by image defacing (all rS > 0.9, P < .0001).
CONCLUSIONS: The defacing model demonstrates efficacy in removing facial regions across multiple MRI types and exhibits minimal impact on downstream research usage. A software package with the trained model is freely provided for wider use and further development (pediatric-auto-defacer; https://github.com/d3b-center/pediatric-auto-defacer-public). By offering a solution tailored to pediatric cases and multiple MRI sequences, this defacing tool will expedite research efforts and promote broader adoption of data sharing practices within the neuroscience community.
ABBREVIATIONS:
- AI
- artificial intelligence
- CBTN
- Children’s Brain Tumor Network
- CE
- contrast-enhanced
- CHOP
- Children’s Hospital of Philadelphia
- CSA
- cross-sectional area
- LH
- left hemisphere
- NIH
- National Institutes of Health
- RH
- right hemisphere
- SEM
- standard error of the mean
- SLIP
- Scans with Limited Imaging Pathology
- T1W
- T1-weighted
- T2W
- T2-weighted
- TMT
- temporalis muscle thickness
SUMMARY
PREVIOUS LITERATURE:
Scientific data sharing promotes reproducibility of research and translation of findings into clinical care. Several centralized repositories have enabled broad sharing of large-scale imaging data sets; however, pediatric data sets have lagged behind their adult counterparts, and neuroimaging data are particularly challenging to share due to privacy concerns, because brain scans can reveal identifiable features. Existing “defacing” tools to remove face regions are primarily designed for adult scans, and often struggle with pediatric images and do not generalize to a variety of sequence types. This work introduces the first tool (pediatric-auto-defacer) specifically for removing facial features from multiparametric pediatric MRIs, addressing a critical gap in data sharing for neuroscience research.
KEY FINDINGS:
A model was developed to automatically remove facial regions from brain MRIs for anonymization purposes. It performs well on several sequence types across various acquisition parameters, and does not over-remove brain tissue. Based on testing, defacing does not affect downstream analytical pipelines (eg, image preprocessing or measured group-level trends).
KNOWLEDGE ADVANCEMENT:
To facilitate broad sharing of pediatric neuroimaging data sets, a robust, automatic deidentification tool is provided to ease the burden on research teams to prepare and release imaging data while protecting patient privacy.
Data sharing is a critical component of research endeavors as it lends to scientific transparency and data reuse. For the study of rare diseases, data sharing is crucial for gathering a meaningful group of samples to enable statistical comparisons in the given patient population. Due to calls to action across disciplines, data sharing plans have recently become a mandate for National Institutes of Health (NIH)-funded projects and deposit of data files to centralized repositories is now a requirement by many scientific journals for publication. Such efforts will facilitate the reproducibility of research studies and consequently their translation into real-world applications such as clinical care contexts, as well as bolster the inclusion of historically under-represented populations, which can mitigate bias in developed models and support fair artificial intelligence (AI) in health care.1
In alignment with FAIR2 principles, several imaging data repositories have been established such as the Alzheimer Disease Neuroimaging Initiative3 and the National Cancer Institute’s The Cancer Imaging Archive4 and Imaging Data Commons, which provide effective data discovery and accessibility. While several large-scale, multi-institutional imaging data sets exist, such as the National Lung Screening Trial (NLST) for lung cancer (chest CTs from more than 26,000 patients)5 and the Breast Cancer Screening Digital Breast Tomosynthesis (breast mammograms from 5060 patients),6 comparable radiology data sets in neuroscience fields have lagged behind their counterparts, primarily due to greater difficulty of removing identifying information from brain (head and neck) scans. Brain images can be inherently identifiable due to the presence of an individual’s face, and their release can jeopardize patient privacy. Studies have shown brain MRIs can be used to identify subjects by matching to their photograph,7,8 even after face regions have been blurred.9 “Defacing,” or the removal of face regions in an image, is one way to mitigate this issue, and several defacing software tools for structural brain MRIs have been developed (eg, mri_deface10, pydeface11, fsl_deface,12 and others13,14), some of which have less impact on downstream processing than others.15,16 That said, existing tools do not typically perform well on pediatric cases,17 particularly in young children and infants, likely due to differences in brain and face anatomy across developmental stages. For example, 1 study found that FSL’s defacing removed brain tissue in most children (ages 8–11) and in some young adult (ages 19–31) cases, and had worse performance for eyes and mouth removal compared with adults.18 FreeSurfer had better performance for face removal without impacting brain tissue in the same cases, however, it was more invasive in removing intraorbital and brainstem structures. Many tools rely on alignment to standardized face or brain atlases created with adult MRIs, and therefore fail to properly deface pediatric scans. Additionally, most are developed for T1-weighted (T1W) sequences, and there remains a need for accessible tools for defacing additional sequence types collected under standard clinical imaging protocols (eg, T2-weighted [T2W]).
Pediatric data sharing has been significantly hindered by regulatory barriers related to privacy concerns, creating a critical unmet need for public imaging data sets. Herein, we build a tool to enable automatic removal of face regions from multiple types of pediatric MRIs, with the goal of facilitating data sharing across neuroscience fields. This is, to the best of our knowledge, the first available pediatric defacing tool. To address the need for a tool that can operate across multiparametric MRIs, we use a large, multi-institutional clinical radiology data set (Children’s Brain Tumor Network [CBTN]19) with deep learning AI methods to develop a model for minimally invasive defacing. Our model was trained and validated with 208 pediatric brain tumor subjects (832 total images) and 36 clinical control subjects (144 images from the Scans with Limited Imaging Pathology [SLIP] cohort20), with 4 image sequences included per subject (T1W, T1W contrast-enhanced [T1W-CE], T2W, and T2W-FLAIR sequences). Images were acquired through clinical protocols, and thus capture real-world heterogeneity in scanner and image acquisition properties.
MATERIALS AND METHODS
Patient Cohorts
Retrospective data were collected from the CBTN,19 a large-scale, multi-institutional repository of longitudinal clinical, imaging, genomic, and other paired data.21 Two hundred eight subjects were selected based on imaging availability and inclusion of a range of ages at the time of imaging (median age 8; minimum = 0.35, maximum = 21.71 years) and cancer histologies (Fig 1, Table, Supplemental Data). MRI scans were unprocessed images from treatment-naïve clinical examinations (T1W, T1W-CE, T2W, and T2W-FLAIR). All subjects had histologically confirmed pediatric brain tumors.
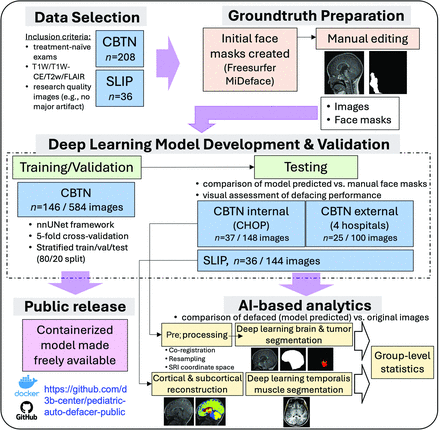
Diagram of overall study workflow. Data cohorts included brain tumor (CBTN) and nonbrain tumor control (SLIP). Initial ground truth face masks were created with MiDeface and manually edited. A 3D deep learning model was trained with the nnUNet framework, by using a single image as input, and tested on withheld data. The impact of defacing on downstream image processing and AI-based pipelines was evaluated with CBTN and SLIP testing data. The trained model is provided in an open-source software container on GitHub.
Patient characteristics in the studied cohorts
To test generalizability to nonbrain tumor patients (clinical control group), a cohort of 40 subjects with available images from the SLIP20 data set were selected to match the general distributions of age and sex of the CBTN cohort. Thirty-six subjects had sufficient images and were included in the main analyses.
Ground Truth Creation with Semiautomated Face Mask Segmentation
Preliminary face masks were generated for each image by using the MiDeface22 algorithm and then were manually edited. Of the 976 images, 507 (52%) were found to be inaccurately defaced and were manually revised by using the ITK-SNAP23 software (by authors C.S., E.G.; Supplemental Data). The criteria for an accurate face mask was that any brain region or temporalis muscle (given potential implications as a biomarker24) were not affected and identifiable facial features, including eyes, nose, mouth, and ears were fully included. Common corrections included restoring brain voxels, particularly in the right prefrontal cortex, and properly realigning the face mask to the subject’s face.
AI Deep Learning Model Development
CBTN images were stratified into training/validation and testing sets (80–20 split) based on demographics (age, sex, race) and histology (Table). nnUNet25 v1 (https://github.com/MIC-DKFZ/nnUNet/tree/nnunetv1; 3D full resolution; Supplemental Data) was used with 5-fold cross-validation, initial learning rate 0.01, stochastic gradient descent with Nesterov momentum (μ = 0.99), and number of epochs = 1000 × 250 minibatches. Each unprocessed T1W/T1W-CE/T2W/FLAIR sequence was treated as a separate input. The set of 4 images for each subject could be used for either training or validation but not both (ie, images from a single subject could not be split into training and validation within a given fold). Given a large percentage of the CBTN scans were from Children’s Hospital of Philadelphia (CHOP), we additionally split the testing cohort into “internal” (CHOP) and “external” (4 separate institutions) testing data sets.
Defacing Accuracy
Model performance was evaluated with (previously unseen) images in the testing cohorts. Traditional performance scores such as the Sørensen-Dice score (spatial overlap between model predicted mask and ground truth mask), sensitivity (percent of pixels correctly identified by the model), and 95% Hausdorff distance metrics (distances between nearest voxels in the predicted and ground truth masks, of which 95% of voxels fell within) were generated.
As an additional assessment of defacing accuracy, 2 raters (authors Neda K. and Nastaran K.) evaluated model performance in the testing cohorts. For each image, they rated coverage of the eyes and ears (separately for left and right), mouth, and nose with either: 1 (fully covered), 0.75 (approximately 75% masked), 0.5 (50% masked), 0.25 (25% masked), or 0 (not masked at all); and whether any brain tissue was removed (yes/no). After initial independent review, images with disagreement were reviewed until a consensus was reached.
Impact of Defacing on Downstream Analytics
Given the overarching aim to facilitate data sharing of brain MRIs for research purposes, it is essential any modification of the images by defacing minimally impacts downstream analysis. Several methods were used to assess this by using standard image processing steps, in both the brain tumor (CBTN) and nonbrain tumor (SLIP) groups separately.
Preprocessing and Application of Pretrained AI Models.
For each subject in the CBTN testing cohorts, T1W, T2W, and FLAIR sequence images were coregistered with their corresponding T1W-CE sequence and resampled to an isotropic resolution of 1 mm3 based on the anatomic SRI24 atlas26 by using the Greedy algorithm (https://github.com/pyushkevich/greedy)27 in the Cancer Imaging Phenomics Toolkit open-source software v.1.8.1 (CaPTk, https://www.cbica.upenn.edu/captk).28 Accuracy of coregistration was confirmed by visual assessment of the 4 images.
Preprocessed data for each subject were then input into existing pretrained AI models for automatic brain tissue extraction and tumor subregion segmentation (https://github.com/d3b-center/peds-brain-seg-pipeline-public).29,30 This was performed once by using the original images (nondefaced), and once by using the defaced images. Resulting brain and tumor segmentation masks were compared between these conditions.
Cortical and Subcortical Volumetric Measures.
For 31 subjects in the SLIP testing cohort, their T1W scan was input to FreeSurfer’s reconstruction pipeline (recon-all; https://surfer.nmr.mgh.harvard.edu/fswiki/recon-all)31 to generate cortical and subcortical structure parcellations (5 subjects were excluded due to insufficient T1W image quality). This was performed once with original images and once with defaced images. Resulting volumetric measurements based on the parcellations were compared between these conditions.
We additionally used an existing AI-powered pipeline to estimate the thickness (temporalis muscle thickness [TMT]) and cross-sectional area (CSA) of the temporalis muscle (https://doi.org/10.5281/zenodo.8428986)24 for 28 SLIP subjects (5 subjects excluded for insufficient quality T1W images, 3 subjects excluded for being younger than 3 years of age as required by the tool).
Please see Supplemental Data for a description of all statistical comparisons and a CLAIM checklist to indicate alignment with the proposed methodologic guidelines recommended for AI in medical imaging.32⇓–34
RESULTS
Defacing Accuracy
Across images, Dice scores indicated decent spatial overlap between manual ground truth and model-predicted face masks in the internal (mean = 0.78, median = 0.8, standard error of the mean [SEM] = 0.008), external (mean = 0.75, median = 0.78, SEM = 0.02), and clinical control (mean = 0.75, median = 0.77, SEM = 0.01) groups (Fig 2). Repeated-measures ANOVAs confirmed there was no effect of image type (T1W/T1W-CE/T2W/FLAIR) on Dice scores in the internal (F(3,108) = 0.38, P = .77) and external (F(3,72) = 1.8, P = .16) cohorts, however there was a significant effect in the clinical control group (F(3,105) = 6.14, P = .007) with better model performance for T2W and FLAIR compared with T1W and T1W-CE (Supplemental Data). Pearson correlations showed no effect of age on Dice scores averaged across image types (internal: r(35) = 0.19, P = .25; external: r(23) = 0.29, P = .17; control: r(34) = 0.28, P = .095; Supplemental Data). One-way ANOVAs indicated no effect of sex (internal: F(1,35) = 2.0, P = .17; external: F(1,23) = 0.28, P = .6; control: F(1,34) = 3.17, P = .08) or race (internal: F(3,33) = 0.18, P = .911; external: F(2,22) = 0.61, P = .551; control: F(2,32) = 1.07, P = .356) on Dice scores, and no effect of histopathologic diagnosis (internal: F(4, 32) = 0.442, P = .777; external: F(1, 23) = 0.377, P = .545) or general tumor location (internal: F(4,32) = 0.837, P = .512; external: F(3,21) = 0.1, P = .959) in the CBTN testing cohorts.
Model performance results. Plots show aggregate metrics across image types for each testing cohort (see Supplemental Data for results for image type separately); error bars represent SEM. A, Standard metrics for segmentation evaluation including Dice similarity, sensitivity, and 95% Hausdorff distance. B, Average performance ratings based on visual inspection by 2 raters (1 = fully covered, 0.75 = approximately 75% masked, 0.5 = 50% masked, 0.25 = 25% masked, 0 = not masked at all).
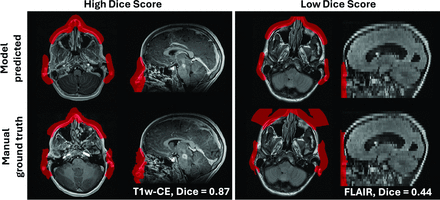
On further review, it was determined that the spatial metrics were not an ideal measure of defacing performance due to variability in extension of the face mask into the air in front of the face in the ground truth segmentations (Fig 3, Supplemental Data). To more accurately assess model performance, 2 raters (Neda K., Nastaran K.) reviewed each defaced image in the internal, external, and clinical control testing groups. After applying the model-predicted face masks to the corresponding images, the raters were instructed to score the model’s accuracy in masking (coverage of) the left eye, right eye, nose, mouth, left ear, and right ear separately (1 = fully masked, 0.75/0.5/0.25 = % partially masked, 0 = not masked) for each image separately.
Representative example images of model predicted versus manual ground truth segmentation masks. Subjects shown with high (left box; T1W-CE sequence) and low (right box; FLAIR sequence) Dice similarity scores between the model predicted (upper row) and manual ground truth (lower row) face masks. This illustrates how Dice score, although a common metric for such segmentation tasks, was not an accurate measure of model performance in the present study, as ground truth masks were variable in their extension into space in front of the face (particularly due to “MiDeface” lettering imposed by the MiDeface Freesurfer tool that was used to generate initial face masks).
Across facial features, the average rated accuracy of model defacing was high for each testing set (means: internal = 0.93, external = 0.86, control = 0.89). Composite scores combining the eyes, mouth, and nose ratings indicated high masking performance for these features (Fig 2, Supplemental Data; internal = 0.97, external = 0.98, control = 0.98), while performance for masking the ears was lower (internal = 0.85, external = 0.62, control = 0.72). For every image, both raters reported no brain voxels were impacted by defacing in the internal, external, or clinical control groups. Repeated-measures ANOVAs showed a significant effect of image type on defacing performance in the clinical control group (F(3,75) = 10.8, P < .0001), with higher average ratings for T1W (M = 0.91) and T1W-CE (M = 0.91) compared with T2W (M = 0.89) and FLAIR (M = 0.86); but no effect of image type in the internal (F(3,108) = 1.17, P = .33) or external (F(3,72) = 0.32, P = .81) groups. Average rating across subjects and image types for each feature is displayed in the Supplemental Data.
Assessing Impact of Defacing on Downstream Analytics
Preprocessing and Application of Pretrained AI Models.
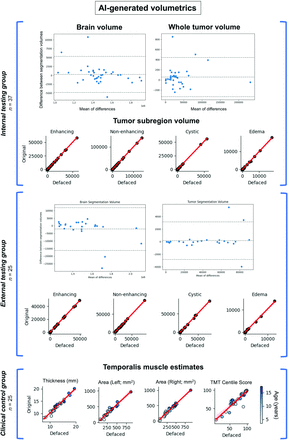
Defaced and original (nondefaced) images underwent preprocessing and were input to pretrained AI tools to assess any impact of defacing on standard downstream analysis by using all 4 image sequences (T1W/T1W-CE/T2W/FLAIR). Visual inspection showed equivalent coregistration performance between defaced and original images. For the pediatric brain tumor test data sets, the volumes of AI-generated brain masks were equivalent between defaced and nondefaced images (internal: rS(35) > 0.99, P < .0001; external: rS(23) > 0.99, P < .0001; Fig 4, upper and middle). AI-generated tumor segmentations were also unaffected by defacing, indicated by equivalent volumes of contrast-enhancing tumor, nonenhancing tumor, cystic, and edema subregions (internal: all subregions rS(35) > 0.99, P < .0001; external: all subregions rS(23) > 0.99, P < .0001; Fig 4, Supplemental Data).
Testing the impact of defacing on AI-generated volumetrics. Each point represents 1 subject; the red line indicates a linear trend. Upper/middle: Comparison of tumor subregion volumes between defaced (x-axis) and original (y-axis) images in pediatric brain tumor subjects. There was very high agreement between brain and tumor segmentation volumes. Lower: Comparison of estimated TMT, area (CSA), and TMT centile scores between defaced (x-axis) and original (y-axis) T1W images from the clinical control group (point colors indicate age). Correlations indicated very high agreement between TMT, CSA, and resulting TMT centile scores.
Cortical and Subcortical Volumetric Measures.
For 31 subjects in the clinical control (SLIP) cohort, we further investigated any impact of defacing on derived brain measures from T1W images by using a standard anatomic reconstruction pipeline (FreeSurfer recon-all). There was very high agreement between estimated global and regional measures, with all comparisons between original and defaced images being positively significant (mean rS(29) = 0.93, all P < .0001; Supplemental Data). Correlations were above 0.9 for 48 out of 58 measures. Regions with the lowest agreement were the left and right cerebellum white matter (left: rS(29) = 0.71, P < .0001; right: rS(29) = 0.69, P < .0001). Nine global measurements (cortex, cerebral white matter, subcortical gray matter, total gray matter, total brain [including cerebellum], total brain excluding ventricles [surface], total brain excluding ventricles [volume], CSF, and total intracranial volumes) were equivalent between original and defaced (rS(29) > 0.86). Paired t tests indicated no significant differences between original and defaced brain measures (Supplemental Data), with the exception of the right vessel (original = 11.3, SEM = 1.38; defaced M = 14.7, SEM = 2.19; t(30) = −2.32, P = .03) and the right hippocampus (original M = 3940.8, SEM = 101; defaced M = 3972.8, SEM = 101; t(30) = −2.36, P = .03), which were estimated to be slightly larger on average in the defaced compared with original images. Overall, these results indicate defacing had minimal impact on cortical and subcortical volumetric assessments by using a standard processing pipeline, which aligns with previous report of minimal effects of defacing tools on global FreeSurfer measurements.17
To examine the impact of defacing on regional measurements in close proximity to the face, we extracted TMT (mm) and CSA measurements (SLIP cohort ages >3 years; n=28) by using an existing AI-powered pipeline24 with T1W images. Notably, TMT scores have been implicated as a predictive marker for sarcopenia across patient populations.35⇓⇓–38 Spearman correlations showed high agreement of estimated TMT (rS26) = 0.96, all P < .0001) and CSA (left hemisphere [LH]: rS26) = 0.96, P < .0001; right hemisphere [RH]: rS26) = 0.97, P < .0001; Fig 4, lower) between defaced and original images. Paired t tests indicated no difference in TMT volumes between original and defaced images (t(27) = −1.8, P = .08), but a significant difference in CSA (LH: t(27) = −3.74, P < .0001; RH: t(27) = −4.79, P = .0009) with lower surface area estimates for the defaced (LH: M = 306.2, SEM = 30; RH: M = 314.7, SEM = 33) compared with original (LH: M = 339.9, SEM = 35; RH: M = 350.5, SEM = 37) images. Resulting centile scores based on TMT, age, and sex (compared with TMT distributions estimated from large-scale data sets24) were not significantly affected by defacing (rS(26) = 0.9, P < .0001; t(27) = −0.97, P = .34).
DISCUSSION
Data sharing of MRIs is crucial to transparent and reproducible research, particularly in the era of predictive AI that requires ample volumes of representative data. Widely available pediatric imaging data sets are needed to accelerate discoveries in neuroscience, particularly in rare disease contexts. To this end, we aim to enable MRI data sharing through the development of an open-source de-identification tool for the automatic removal of identifiable facial features. A deep learning model for face masking was trained by using a large, multi-institutional data set of clinically acquired, multiparametric MRIs (CBTN).
The trained model had strong performance removing the face (eyes, nose, mouth) in an unseen data set, with adequate, though lower, performance on ear removal. This is potentially due to a lack of presence of ears in some images in the training data set (limited field of view). Notably, although the model was trained on data from patients with brain tumor, it could generalize to a separate data set of clinically matched controls indicating its potential use across anatomically normal and disease-impacted cohorts. To enable wider usage by the community, the trained model is publicly provided as an open-source software package, and we encourage further model development to extend the model to additional disease and healthy populations (see potential clinical limitations in the Supplemental Data).
Critically, image alteration by defacing should not impact usage in intended research purposes. To ensure this, we compared the outputs of standard processing pipelines between defaced and original (nondefaced) images. Statistical trends for AI-estimated whole brain and tumor volumes (brain tumor group), in addition to derived brain region volumes, global brain metrics, and AI-generated temporalis muscle measurements (control group), were unaffected by defacing. Most estimated measures were equivalent between defaced and original images, and any resulting measurement differences did not impact overall patterns at a group-level. Thus, there was minimal impact of defacing on the utility of the structural images for downstream analysis with standard research pipelines.
Many existing defacing tools are limited to T1W sequences,13,22,39 and we sought to expand support to additional structural image types (T2W, FLAIR, T1W-CE), given their prevalence in clinical and research practices. That said, our tool is limited to 4 sequences, and further development could expand to additional types such as functional MRI and other advanced imaging (eg, diffusion-weighted imaging). Although consensus review was used to assess defacing performance, additional quantitative metrics such as face recognition rate may provide a more objective measure of de-identification performance. Another limitation of this study is that, while the training data set included images across 6 institutions, a large portion of the data set came from a single institution (CHOP). Future work should focus on expanding to larger studies to bolster model generalizability, and would benefit from direct comparison between deep learning and existing computer-vision methods.
CONCLUSIONS
We developed an AI-powered pediatric defacing tool with the goal of facilitating wider de-identification of structural MRIs for data sharing purposes. The tool is publicly available (https://github.com/d3b-center/pediatric-auto-defacer-public) and can be used on multiple image types. Future work can extend the model to additional populations and MR sequences to provide a universal method to facilitate data sharing and ultimately drive discoveries in neuroscience research.
Footnotes
This project was supported in part from the National Institutes of Health (NIH) National Heart, Lung, and Blood Institute (NHLBI; grant number U2CHL156291/3U2CHL156291-02S1 to A.C.R.).
Disclosure forms provided by the authors are available with the full text and PDF of this article at www.ajnr.org.
References
- Received July 24, 2024.
- Accepted after revision November 7, 2024.
- © 2025 by American Journal of Neuroradiology