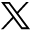Graphical Abstract
Abstract
BACKGROUND AND PURPOSE: Recent advances in deep learning have shown promising results in medical image analysis and segmentation. However, most brain MRI segmentation models are limited by the size of their data sets and/or the number of structures they can identify. This study evaluates the performance of 6 advanced deep learning models in segmenting 122 brain structures from T1-weighted MRI scans, aiming to identify the most effective model for clinical and research applications.
MATERIALS AND METHODS: A total of 1510 T1-weighted MRIs were used to compare 6 deep learning models for the segmentation of 122 distinct gray matter structures: nnU-Net, SegResNet, SwinUNETR, UNETR, U-Mamba_BOT, and U-Mamba_ Enc. Each model was rigorously tested for accuracy by using the dice similarity coefficient (DSC) and the 95th percentile Hausdorff distance (HD95). Additionally, the volume of each structure was calculated and compared between normal controls (NCs) and patients with Alzheimer disease (AD).
RESULTS: U-Mamba_Bot achieved the highest performance with a median DSC of 0.9112 (interquartile range [IQR]: 0.8957, 0.9250). nnU-Net achieved a median DSC of 0.9027 [IQR: 0.8847, 0.9205], and had the highest HD95 of 1.392 [IQR: 1.174, 2.029]. The value of each HD95 (<3 mm) indicates its superior capability in capturing detailed brain structures accurately. Following segmentation, volume calculations were performed, and the resultant volumes of NCs and patients with AD were compared. The volume changes observed in 13 brain substructures were all consistent with those reported in existing literature, reinforcing the reliability of the segmentation outputs.
CONCLUSIONS: This study underscores the efficacy of U-Mamba_Bot as a robust tool for detailed brain structure segmentation in T1-weighted MRI scans. The congruence of our volumetric analysis with the literature further validates the potential of advanced deep learning models to enhance the understanding of neurodegenerative diseases such as AD. Future research should consider larger data sets to validate these findings further and explore the applicability of these models in other neurologic conditions.
ABBREVIATIONS:
- AD
- Alzheimer disease
- ADNI
- Alzheimer’s Disease Neuroimaging Initiative
- CNN
- convolutional neural network
- DSC
- dice similarity coefficient
- HD95
- 95th percentile Hausdorff distance
- IQR
- interquartile range
- NC
- normal control
- SSM
- state-space sequence model
SUMMARY
PREVIOUS LITERATURE:
Previous studies have demonstrated the utility of CNNs and hybrid transformer models in medical image segmentation, particularly in neuroimaging. U-Net–based architectures have been widely adopted for their ability to capture spatial details, while transformer models show promise in capturing global dependencies. However, many approaches still face limitations, such as computational resource demands and high memory usage, when applied to large-scale data sets. The introduction of structured state-space models, such as U-Mamba, provides a new perspective on improving both segmentation accuracy and computational efficiency in biomedical imaging.
KEY FINDINGS:
Our study found that U-Mamba_Bot outperformed other models, achieving the highest DSC of 0.9112. It also exhibited competitive training and inference times compared with other architectures.
KNOWLEDGE ADVANCEMENT:
The U-Mamba model’s integration of structured state-space mechanisms addresses some of the limitations of traditional CNN and transformer-based models, particularly in capturing long-range dependencies with lower computational cost. These findings highlight U-Mamba’s potential for enhancing neuroimaging analysis in clinical applications.
MRI can deliver superior spatial and contrast resolution and has become a cornerstone in diagnosing and treating neurologic disease. Instance segmentation refers to delineating intracranial structures (segmentation) and assigning individual labels to every structure, which is essential in studying brain MRI. It provides valuable information for structural analysis, volumetric assessment, surgical planning, and image-guided intervention. For example, several studies from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) demonstrated that alterations in intracranial structural volumes (as quantified by using segmentation tools) correlate with outcome measures in clinical trials for Alzheimer disease (AD). A study from Radue et al1 demonstrated that brain volume loss correlated with clinical and radiologic outcomes in patients with multiple sclerosis, and Mora et al2 demonstrated that patients with medial temporal lobe epilepsy exhibit a consistent pattern of gray matter atrophy on MRI, suggesting that a common pathophysiologic process may be responsible for the disease. Thus, diagnosing neurologic and neuropsychiatric diseases necessitates a comprehensive understanding of subcortical structures, and it is crucial to grasp both the structural and functional characteristics of the brain.
Automated segmentation methods have been developed to differentiate between frontotemporal dementia and AD based on MRI.3 Recent advances in deep learning have shown promising results in medical image analysis and segmentation.4 State-of-the-art methods, such as U-Net and transformers, have achieved impressive success in medical image segmentation.5 The convolutional neural network (CNN)-based U-Net architecture has been widely utilized in various fields, particularly medical image segmentation, due to its effectiveness in capturing spatial information and features. Though initially developed for natural language processing tasks, transformers have recently been adapted for medical image segmentation and offer promising results.6,7 Though U-Net can better recognize local features and transformers can capture global features, given their respective advantages and disadvantages, many current approaches are combining the two, for example, UNETR8 and SwinUNETR9 for more accurate results and more robust performance. However, certain shortcomings still exist, such as being resource-intensive, and having high memory and computational requirements.
Recently, state-space sequence models (SSMs), particularly structured SSMs, have emerged as efficient and powerful components for constructing deep networks that deliver top-tier performance in continuous long-sequence data analysis.10 Mamba improved the structured state-space sequence models model by introducing a selective mechanism that allows the model to select relevant information depending on the input.11 U-Mamba was newly developed for general purpose biomedical image segmentation with a self-adapting function network based on the innovative hybrid CNN-SSM architecture. Compared with the currently popular deep learning network architectures, nnU-Net, SegResNet,12 and transformer-based SwinUNETR,9 U-Mamba achieved the best results in image segmentation for abdominal MRI, instruments in endoscopy, and cells in microscopy.13
In this study, we compared the newly released U-Mamba automatic segmentation model and previous state-of-the-art segmentation models for whole-brain substructure segmentation on T1-weighted MRI. We utilized the ADNI database to conduct comparative analyses of the most popular current models for medical image segmentation for almost all cortical subregions and nuclei of the human brain. This sets the stage for further understanding the relationship between brain structural changes and diseases, uncovering unknown disease mechanisms, and providing highly automated and robust tools. We believe these have significant potential for clinical application.
MATERIALS AND METHODS
Data Collection
Data used in the preparation of this article were obtained from the ADNI database (http://adni.loni.usc.edu). The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, PET, other biologic markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment and early AD.
This study analyzed 20,056 randomly selected T1-weighted MRIs from the ADNI data sets. Because each patient typically undergoes multiple MRI scans, we retained the initial MRI from each patient in the ADNI database. We excluded cases lacking detailed information and ultimately obtained 1510 MRI scans for this study (Table 1). Normally distributed data are expressed as means (standard deviation), and non-normally distributed data are expressed as medians with interquartile range (median [IQR: 25th, 75th]).
Demographics of the data set obtained for the study
Data Labeling and Preprocessing
To generate ground truth labels (pixel-level segmentation masks), we linearly registered the Mayo Clinic Adult Lifespan Template and Atlas to every volume by using ANTs tool (https://github.com/ANTsX/ANTs) resulting in 122 gray matter structures.14,34 Board-certified radiologists visually inspected all volumes to ensure good image quality and proper registration. We divided the data set into 768 training, 192 validations, and 550 for testing (all from different patients) for model evaluation. Table 1 summarizes the demographics and characteristics of the patients in this data set.
Because our study is focused on brain segmentation, the deep learning model requires the removal of nonbrain tissue to increase the model’s performance. A deep learning-based brain extraction tool, HD-BET,15 was used on T1-weighted MR images.
Model Training
All of the deep learning models were adapted to work within the nnU-Net framework. nnU-Net is a deep learning-based segmentation method that automatically configures and runs the entire segmentation pipeline for any biomedical image data set, including preprocessing, data augmentation, model training, and postprocessing. The pipeline handles hyperparameter tuning and does not require any changes to the network architecture. Therefore, it provides a perfect environment for comparing U-Mamba with other methods. Also, it enables U-Mamba to be easily adapted to a wide range of segmentation tasks. In the nnU-Net framework, the patch size is 128 × 128 × 128 with a batch size equal to 2. The Adam optimizer with an initial learning rate of 0.01 was used to optimize network weights, with the momentum set at 0.99. An empirical combination of Dice loss with cross-entropy loss in nnU-Net has enhanced training stability and improved segmentation accuracy.13,16 To ensure a fair comparison, we also implemented SegResNet,12 UNETR,8 and SwinUNETR9 into the nnU-Net framework and utilized nnU-Net recommended and default optimizers for model training.13 For more detailed information on how each model was adapted within the nnU-Net framework, please refer to the GitHub repository: https://github.com/wyjzll/U-Mamba. This repository includes comprehensive documentation and code examples illustrating the integration process. To ensure the reproducibility of this study and maintain consistency in the code version used, independent of updates by the original U-Mamba authors,13 we forked the original U-Mamba GitHub repository (https://github.com/bowang-lab/U-Mamba). This approach guarantees that the code remains unchanged, allowing others to reliably replicate our findings. All models were trained on 1 graphics processing unit (GPU) (NVIDIA A100 80G SXM) for 1000 epochs with random initial weights. The entire process of this study is presented in Fig 1.
An overview of the study flow. A, T1-weighted MR images were first processed through a brain extraction step by using the brain extraction tool-HD-BET. B, The processed image then serves as the input to each of the deep learning models, which are responsible for segmenting the brain into different anatomic regions. C, The evaluation process was conducted by comparing the segmentation results for each model with the ground truth.
Evaluation
The performance of the models in segmenting all 122 brain substructures was assessed by using the dice similarity coefficient (DSC) and the 95th percentile Hausdorff distance (HD95).
The DSC is defined in equation 1:
 (1)
(1)
Here, X and Y represent the ground truth segmentation and model segmentation. The ∣⋅∣ indicates the number of elements in the set and ∩ represents the intersection of the sets.
The HD is defined in equation 2:
 (2) where:
(2) where:
A and B are the 2 sets of points (eg, the edges of the segmented regions for the ground truth and for a model)
h(A, B) is defined as maxa ∈ A minb ∈ B d(a, b)
h(B, A) is defined as maxb ∈B mina ∈A d(b, a)
d(a, b) is the distance between points a and b (typically Euclidean distance).
The HD95 is calculated by taking the 95th percentile of all the computed distances rather than the maximum, helping to ignore the most extreme values that might be due to noise or other anomalies. This makes it a robust measure for assessing the accuracy of medical image segmentations, where outliers can skew the results.
Statistics
We use the Kolmogorov-Smirnov test for all normality tests for all of the groups. Two-sample t tests were used to compare differences in normally distributed data between groups. If data were not normally distributed, the data were tested by using nonparametric approaches (Mann-Whitney U test). The statistical tests were performed by using GraphPad Prism 10.0 and SciPy 1.8.0. Results with P < .05 were indicated by asterisks.
This article follows the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis Or Diagnosis Checklist.35
RESULTS
Model Performance
A representative mask generated by our model is shown in Fig 2. All 122 gray matter regions were segmented successfully in all of the models (that is, no structure had a DSC of 0 for any model (Supplemental Data). We have provided downloadable result samples on our GitHub page (https://github.com/wyjzll/Brain_Segmentation) for interested readers to further reference. The models evaluated were nnU-Net, SegResNet, UNETR, SwinUNETR, U-Mamba_BOT, and U-Mamba_Enc. Each model’s effectiveness is assessed by using the DSC and HD95. A success rates of DSC > 0.9 or HD95 < 3 mm was defined to ensure high accuracy and reliability in clinical settings (Table 2). Among these models, U-Mamba_Bot showed superior segmentation accuracy with the highest DSC value of 0.9112 [IQR: 0.8957, 0.9250], achieving high success rates of 68.85% for DSC > 0.9, this model, however, did not have the best HD95 score, suggesting a potential trade-off between general overlap and boundary precision. U-Mamba_Enc demonstrated competitive performance, closely matching nnU-Net in DSC (0.8968 [IQR: 0.8801, 0.9155]), but with a higher HD95 of 1.544 [IQR: 1.224, 2.318]. In contrast, the SegResNet, while having a competitive DSC score (0.9033), exhibited a median HD95 (1.449 mm), which could imply higher boundary accuracy compared with other models. The UNETR model, as presented in the chart, showed a DSC of 0.8709 with an IQR from 0.8521 to 0.8978, which was the lowest among the models tested. This suggests that it may have some limitations in achieving a high degree of overlap between the predicted and true segmentations compared with the other models.
Representative images generated by the deep learning models versus human-labeled ground truth.
Results summary of trained segmentation models on T1-weighted MRI data sets
As shown in Table 3, the training times varied significantly among models. nnU-Net V2 has the longest epoch time at 412 seconds, indicating that it may require considerable computational resources and time for training. U-Mamba_Enc had the fastest training time, completing an epoch in just 169 seconds. SegResNet and UNETR also showed relatively quick epoch times at 183 and 191 seconds, respectively, making them suitable for environments in which faster model training is beneficial. SwinUNETR and U-Mamba_Bot fell in the middle, with epoch times of 314 and 273 seconds, respectively, balancing computational complexity and training speed.
A comparison of the epoch training times and inference time per image for different deep learning models
To assess the effectiveness of the models, we calculated the degree of correspondence between the volumes of individual brain regions in each model’s predictions and the ground truth. Figure 3 shows the performance of various deep learning models in segmenting brain substructures. Each model’s accuracy was evaluated based on its ability to match ground truth measurements, with the results categorized into significant differences (P < .05) and nonsignificant differences (P > .05). The models included in the analysis are nnU-Net, SegResNet, UNETR, SwinUNETR, U-Mamba_Bot, and U-Mamba_Enc. SegResNet and U-Mamba_Bot aligned more closely to the ground truth, indicated by more nonsignificant differences versus ground truth (117 and 118, respectively). nnU-Net demonstrated a high discrepancy from ground truth with 122 significant differences.
Comparison of the number of brain substructure volumes that had significant differences from ground truth by using different models.
Clinical Testing
Patients with AD are intricately linked to changes in brain structure and volume characterized by brain atrophy. Understanding these alterations through neuroimaging studies is crucial for the early detection and monitoring of disease progression. In this part, we analyzed the prediction results produced by U-Mamba_Bot on the test set, which includes 134 healthy individuals and 112 patients with AD. We found that among all the brain areas that atrophied in patients with AD, the amygdala exhibited the most significant volume reduction compared with the NC group, with the left and right sides shrinking by 13.03% and 10.03%, respectively. This was followed by the bilateral entorhinal cortex, which decreased by 8.60% on the left side and 9.33% on the right. In contrast, the volumes of the caudate in the NC and AD groups increased by 8.74% and 7.27%, respectively, which aligns with the findings reported in the literature.17 For ease of presentation, we only show the volume ranges of the 13 brain regions that have statistically significant differences between the NC and AD groups (Fig 4), based on the image segmentation results generated by the U-Mamba_Bot model.
Comparison of brain substructure volumes between NC and AD. The dual-colored bars represent quantified volumes for each respective brain substructure, highlighting significant discrepancies between the 2 groups (*P < .05, Mann-Whitney U test).
DISCUSSION
In this paper, we have chosen the latest U-Mamba model to conduct what we believe to be the most extensive substructural segmentation of the brain to date. Our comprehensive experiment effectively highlights the variability in the accuracy of most deep learning models in replicating precise brain substructure volumes, providing insights into their reliability for clinical and research applications, and suggested model selection based on specific requirements for accuracy and margin precision in future clinical applications. The results indicate that U-Mamba’s performance surpasses that of existing CNN and Transformer-based segmentation networks across different modes and segmentation targets. In particular, U-Mamba has significantly faster training speeds compared with CNN and Transformer architectures. This is instrumental in addressing challenges posed by the local nature of CNNs and the computational complexity of Transformers, which affect long-range modeling. This advantage is largely attributed to the architectural design of U-Mamba, which is capable of extracting multiscale local features while capturing long-range dependencies.
The nnU-Net framework, based on the U-Net architecture, has demonstrated exceptional performance in various segmentation tasks, surpassing state-of-the-art models in international biomedical image segmentation challenges. Its success can also be attributed to adaptive preprocessing, extensive data augmentation, model ensembling, and aggregating tiled predictions, which collectively contribute to its consistently high performance across a wide range of tasks.16,18 nnU-Net can be configured to execute the entire segmentation pipeline automatically; in addition, it offers a range of features that make it highly adaptable and effective across different models and tasks. No other specific data preprocessing (beyond brain extraction) is needed in this part.16 We have chosen the nnU-Net as our segmentation network backbone, which enabled us to focus on implementing the network while managing other variables like image preprocessing and data augmentation. This arrangement facilitates a fair comparison of U-Mamba with other methods under consistent conditions, where the network architecture is the sole variable that differs.
In evaluating the efficiency of different deep learning models, it is essential to consider the training times, which can vary significantly among models. Our results demonstrated that U-Mamba_Enc has the fastest training time among the most popular models. However, U-Mamba_Bot showed the advantage of having the quickest inference time among all the models we evaluated. Based on studies by Gu et al11 and Ma et al,13 Mamba advances SSMs in discrete data modeling, such as text and genomes, through 2 significant enhancements. First, Mamba introduces an input-dependent selection mechanism, a departure from the traditional time- and input-invariant SSMs, enabling effective filtration of information from inputs. This mechanism is achieved by parameterizing the SSM parameters according to the input data.11,13 Second, Mamba incorporates a hardware-aware algorithm that scales linearly with sequence length and computes the model recurrently with a scan, thereby enhancing processing speed on modern hardware.
The Mamba architecture, which combines SSM blocks with linear layers, is notably simpler and has achieved state-of-the-art performance in various long-sequence domains such as language and genomics. This simplicity translates into significant computational efficiencies in the training and inference phases.11,13 Wu et al19 explored the core features of Mamba and conceptually determined that it is best suited for tasks involving long sequences and autoregressive features. For vision tasks that do not have these characteristics, such as image classification, they argue that Mamba may not be necessary. However, while detection and segmentation tasks are not autoregressive, they do involve long sequences.19 Interestingly, our study also shows that the overall performance of U-Mamba_Bot with U-Mamba block applied only at bottleneck achieves the highest DSC value among all models, exceeding that of U-Mamba_Enc with U-Mamba block applied in all encoder parts. This suggests that replacing all encoder modules with SSM blocks may not necessarily yield optimal accuracy. Therefore, it is worth investigating the potential benefits of applying Mamba to such tasks.
Understanding regional brain volume is crucial for comprehending the pathophysiology of various brain-related diseases. Several studies have investigated the relationship between brain volume and different health issues such as Huntington disease,20 atrial fibrillation,21 AD, critical illnesses,22 multiple sclerosis23 Parkinson disease,24 and migraine.25 This study utilized the ADNI database and successfully completed whole-brain substructure image segmentation, followed by the calculation of brain substructure volumes and a comparative analysis with the NC group. We identified 13 brain functional areas (Fig 4) with significant changes in brain volume, most of which align with findings reported in the literature.17,26⇓⇓⇓⇓⇓–32 However, our results did not show the significant reduction in hippocampal volume that other studies have reported,33 and this discrepancy merits further investigation. In future studies, we also plan to apply our study to a larger database and different diseases to offer valuable insights for the diagnosis, prognosis, and monitoring of treatment in different neurologic conditions.
Limitations
We have found that certain types of intracranial lesions, such as displacement, space-occupying lesions, inflammation, trauma, edema, hemorrhage, etc, can significantly impact the performance of our model. Because the ADNI database primarily consists of imaging data from elderly individuals, including healthy controls and those with various cognitive impairments, even though these subjects may exhibit certain pathologic or age-related changes in brain structure, their basic structure remains relatively unchanged. Therefore, our model is not optimized for segmentation tasks involving such structural alterations.
Moreover, we have identified some minor hand-labeling mistakes during the process. For example, CSF signals adjacent to gyri were occasionally mislabeled as gyri. Although these errors did not significantly affect larger brain regions, they could lead to substantial calculation errors in small brain structures. This is an area that requires improvement for future work.
CONCLUSIONS
We successfully segmented gray matter regions by using several popular deep learning models, including U-Mamba, a newly developed deep learning architecture. Our extensive experimental findings demonstrate that U-Mamba’s performance matches that of existing CNN and Transformer-based segmentation networks across various modes and segmentation targets. Specifically, U-Mamba_Bot exhibits a marked increase in accuracy for segmentation models compared with CNN and Transformer architectures. Furthermore, the variability across different brain regions captured in this study not only reinforces the heterogeneity of Alzheimer pathology but also may guide targeted research into the development of diagnostic markers and therapeutic strategies focused on the most affected areas. We believe it may be used clinically for feature extraction, morphologic analysis, and downstream diagnostic tools, thereby contributing to the development of automated diagnostic and treatment assessment systems.
Acknowledgments
We acknowledge all the authors of the employed public data sets, allowing the community to use these valuable resources for research purposes. We also thank the authors of nnU-Net (https://github.com/MIC-DKFZ/nnUNet) and U-Mamba (https://github.com/bowang-lab/U-Mamba) for making their valuable code publicly available.
Data collection and sharing for the Alzheimer Disease Neuroimaging Initiative is funded by the National Institute on Aging (National Institutes of Health Grant U19 AG024904). The grantee organization is the Northern California Institute for Research and Education. In the past, ADNI has also received funding from the National Institute of Biomedical Imaging and Bioengineering, the Canadian Institutes of Health Research, and private sector contributions through the Foundation for the National Institutes of Health including generous contributions from the following: AbbVie; Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics.
Footnotes
Disclosure forms provided by the authors are available with the full text and PDF of this article at www.ajnr.org.
References
- Received June 11, 2024.
- Accepted after revision October 8, 2024.
- © 2025 by American Journal of Neuroradiology