Abstract
BACKGROUND AND PURPOSE: This retrospective study evaluated the utility of contrast-enhanced T1-weighted 3D fast spin-echo–based sampling perfection with application-optimized contrasts by using different flip angle evolutions (SPACE) sequences for brain metastasis detection on 3T MRI compared with a gradient-recalled echo–based 3D FLASH sequence.
MATERIALS AND METHODS: We identified all patients at a single institution who underwent SPACE and 3D FLASH sequences as part of a practice quality-improvement project. Their medical records were retrospectively reviewed. Five certified neuroradiologists reviewed the images, with at least 2 weeks’ separation between scoring sequences for the same patient. We evaluated the following parameters: number of metastatic lesions, number of indeterminate lesions, lesion margin, contrast-to-noise ratio (CNR), extent of image artifacts, and overall image quality. The CNR was also quantified for solidly enhancing lesions of >1 cm.
RESULTS: We identified 220 patients who underwent SPACE and 3D FLASH sequences (the order of the sequences was equally distributed). Of these, 79 had brain metastases on imaging, and 7 were excluded; thus, 72 patients were included in the study. Twenty patients were scored by 2 radiologists. Of the 92 evaluations, SPACE detected more lesions than 3D FLASH in 35, while 3D FLASH detected more lesions in 10. More indeterminate lesions were seen on 3D FLASH (n = 27) than on SPACE (n = 9). For the lesion margin, CNR, and overall image quality on a Likert scale, SPACE performed significantly better than 3D FLASH, with fewer image artifacts (P < .00001). Higher quantitative CNRs were found on SPACE than on 3D FLASH images, though this result was not statistically significant (median = 22.9 versus 15.5, respectively, P = .134). There was a high interreader lesion detection concordance with the Krippendorf α ordinals at 0.962 for SPACE, 0.870 for 3D FLASH, and 0.918 for the 2 sequences combined.
CONCLUSIONS: Compared with 3D FLASH, the SPACE sequence detected more metastatic lesions and was rated higher for image quality, lesion margin, and CNR, with fewer artifacts. Importantly, the SPACE sequence resulted in increased reader confidence, with fewer indeterminate lesions detected.
ABBREVIATIONS:
- CE
- contrast-enhanced
- CNR
- contrast-to-noise ratio
- GRAPPA
- generalized autocalibrating partially parallel acquisition
- IR
- inversion recovery
- SE
- spin-echo
- SPACE
- sampling perfection with application-optimized contrasts by using different flip angle evolution
- VIBE
- volumetric interpolated breath-hold examination
Cerebral metastatic disease is the most common CNS malignancy, with >100,000 patients diagnosed with brain metastases in the United States annually; it results in remarkable morbidity and mortality.1,2 Early detection is important, with improved outcomes directly related to lesion size at treatment,3 highlighting the need for the optimization and selection of high-performing MR imaging sequences.
Contrast-enhanced (CE) T1-weighted 3D MRI is the most important imaging technique for the detection and delineation of brain metastases. Several T1-weighted 3D MRI techniques are in use today, all with different advantages and disadvantages. The ability to visualize gadolinium-based contrast agent enhancement is paramount to the detection of brain metastases. However, vascular enhancement may make it difficult to distinguish vessels from metastases. Magnetization-prepared rapid acquisition with gradient echo (MPRAGE) is the most commonly used T1-weighted 3D MR sequence for neuroimaging, because it provides great tissue contrast between gray and white matter, with high spatial resolution. However, GRE-based sequences have been shown to have lower observed contrast-enhancement than spin-echo (SE) sequences.4
T1-weighted sampling perfection with application-optimized contrasts by using different flip angle evolution (SPACE sequence; Siemens),5 a 3D FSE-based sequence, has been found to improve the detection of small metastatic lesions relative to MPRAGE, with similar spatial resolution at 3T.6-8 In addition, both SPACE and volumetric interpolated breath-hold examination a 3D GRE-based sequence, have demonstrated superior conspicuity for brain tumor enhancement compared with MPRAGE.9 VIBE and MPRAGE are both GRE-based 3D sequences without and with inversion recovery (IR) preparation, respectively. The “consensus recommendations for a standardized brain tumor imaging protocol for clinical trials in brain metastases” recommended IR-GRE (e.g., MPRAGE) as the “minimum standard” pulse sequence at both 1.5T and 3T and 3D FSE (eg, SPACE) as the “ideal protocol,” which is best performed at 3T.10
At our institution, we have used CE T1-weighted 3D fast low-angle shot (FLASH) sequence, the classic spoiled GRE-based sequence without IR, for the detection of brain metastasis. The 3D FLASH sequence has a simpler relationship of signal and contrast as a function of T1 and may provide a larger signal difference with gadolinium-based contrast agent enhancement than MPRAGE.11 While previous studies demonstrated that T1-weighted SPACE is superior to MPRAGE for the detection of brain metastatic lesions,6⇓–8 there is limited evidence showing that it is also superior to non–IR-prepared 3D GRE sequences. In this study, we evaluated the utility of the CE T1-weighted SPACE sequence by directly comparing it with the 3D FLASH sequence for the detection of brain metastases at 3T.
MATERIALS AND METHODS
Subjects
This retrospective study was approved by the institutional review board at our institution, and the patient informed consent requirement was waived. We retrospectively reviewed the medical records of all patients at a single institution who underwent brain MRI with CE T1-weighted SPACE and 3D FLASH sequences as part of a practice quality-improvement project from May 3, 2021, to July 2, 2021. Patients were included if brain metastases were identified on imaging. Patients were excluded if they had incomplete or additional evaluations. For patients who had >1 MRI study, only the first one was included. The STrengthening the Reporting of OBservational studies in Epidemiology. (STROBE) methodology was followed, and the checklist is provided as Supplemental Data.
Image Acquisition
All MRI studies were performed as routine brain MRI on three 3T scanners of the same model (Magnetom Skyra; Siemens) with a 20-channel head coil. Our standard of care protocol included precontrast axial 2D DTI, T1 SE, T2 FSE, T2 FLAIR, and T2* GRE and postcontrast axial 2D SE and 3D FLASH, with the administration of 0.1-mmol/kg gadobutrol (Gadavist; Bayer HealthCare). The 3D FLASH sequence was acquired as axial slices (TR/TE = 6.2/2.7 ms, flip angle = 20°, generalized autocalibrating partially parallel acquisition (GRAPPA) factor = 2, in-plane resolution = 0.9 × 0.9 mm, slice thickness = 0.7 mm with 50% resolution, scan time = 4 minutes and 10 seconds), and reformatted to 1-mm slices in the coronal and sagittal planes. The SPACE sequence was acquired as sagittal slices (TR/TE = 600/11 ms, echo-train length = 38, GRAPPA factor = 2 × 2, in-plane resolution = 1 × 1 mm, slice thickness = 1 mm, scan time = 4 minutes and 3 seconds), reformatted to 1-mm slices in the axial and coronal planes. Both 3D FLASH and SPACE sequences were scanned after the postcontrast 2D SE sequence. The order of the 3D FLASH and SPACE acquisitions was balanced among the patients.
Image Analysis
The SPACE and 3D FLASH images were reviewed on a PACS and graded by 5 Certificate of Added Qualification–certified neuroradiologists, each with >5 years of experience. The reviewers scored only 1 of the 2 sequences at a time. Images were reviewed in batches, with at least a 2-week separation between reviewing sequences on the same patient to avoid memory bias. Of all the studies scored, 20 studies were scored by 2 radiologists randomly assigned from the pool of 5 interpreting radiologists. Reviewers were instructed to focus on the axial images and review the coronal and sagittal images as needed.
For each sequence, the numbers of CE metastatic lesions and indeterminate foci of enhancement were recorded. Indeterminate lesions were defined as punctate or linear foci of enhancement that were subjectively determined by the interpreting radiologist as not definitively a result of cerebral metastatic disease and could be interpreted as vascular in nature.
For each sequence, reading efficiency was evaluated by recording the length of time it took each interpreting radiologist to complete the identification of metastatic or indeterminate lesions. We performed a qualitative visual assessment of lesion margin, contrast-to-noise ratio (CNR), overall image quality, and the extent of artifacts using a 5-point Likert scale (5: excellent, 4: good, 3: acceptable, 2: poor, and 1: unacceptable). CNR was defined as the contrast of enhancing lesions relative to the noise in the background. For each radiologist and subject, the difference in each imaging metric was computed. For example, IQi,j,SPACE − IQi,j,3D FLASH is the difference in image quality scored by radiologist j on patient i. Positive differences indicated the superiority of SPACE to 3D FLASH. The barplots of the differences for each imaging metric were plotted, colored by the radiologist.
A quantitative analysis of CNR was performed in patients with solidly enhancing lesions of >1 cm in the greatest axial dimension. An ROI measuring 0.5 cm2 was drawn by the interpreting radiologist over the enhancing lesion, with a second ROI of the same size drawn over the adjacent normal brain parenchyma, taking care to avoid any vascular enhancement and brain parenchyma with edema or signal abnormality. One measurement was made for each of these patients with lesion ROIs, focusing on the area with the highest enhanced signal. The mean signal intensity and SD of each ROI were recorded, and CNR was calculated using the equation:9
 where SIlesion and SIparenchyma represent the mean signal intensity of the lesion and parenchyma ROI, respectively, and SDparenchyma represents the SD of the parenchymal ROI.
where SIlesion and SIparenchyma represent the mean signal intensity of the lesion and parenchyma ROI, respectively, and SDparenchyma represents the SD of the parenchymal ROI.
To further inspect whether there were large lesions missed by either sequence, a neuroradiologist subsequently reviewed studies with a difference in the number of lesions detected on SPACE versus FLASH. The sequences were reviewed side by side, and metastatic lesions were annotated on SPACE and FLASH. Lesions not well-seen on either sequence were measured in the greatest axial dimension, and the number of inconspicuous lesions of >5 mm was counted.
Statistical Analysis
Statistical analyses were performed in R, Version 4 (http://www.r-project.org/) or the SPSS software package, Version 24.0 (IBM). A linear regression analysis was performed to test the correlation between the number of contrast-enhancing metastases detected on SPACE versus 3D FLASH. Paired Wilcoxon tests were used to compute P values to assess the statistical significance of differences in image metrics between the 2 sequences. For images scored by multiple radiologists, ordinal and interval Krippendorf α was used to measure concordance.12
RESULTS
We identified 220 studies with CE T1-weighted SPACE and 3D FLASH sequences acquired from May 3, 2021, to July 2, 2021. Seventy-nine of them were found to have brain metastases at the time of imaging. Seven studies were excluded (2, the second time point of the same patient; 3, one sequence was evaluated twice; 2, one sequence was not evaluated); thus, 72 patients (43 female and 29 male; mean age, 63.9 [SD, 13.0] years) were included in the analysis (see Table 1 for patient demographic and clinical characteristics). Scans of 20 patients were read by 2 radiologists, resulting in a total of 92 evaluations.
Patients’ demographic and clinical characteristicsa
Across the 92 evaluations, the total number of metastases detected was higher for SPACE (832) versus FLASH (723) with the same median number of metastases detected for each sequence (2). Across the 72 patients (when averaging lesion detection numbers between 2 radiologists for the 20 studies read by 2 radiologists), the total number of metastases detected was higher for SPACE (639.5) than FLASH (521.5) with the median the same for both sequences (2). The numbers of metastatic lesions detected on SPACE and 3D FLASH were significantly correlated (R = 0.98, P < .001), as demonstrated in Fig 1A. Overall, the 2 sequences detected the same number of lesions in 47 cases: SPACE detected more lesions than 3D FLASH in 35 cases, while 3D FLASH detected statistically significantly more lesions in 10 (Fig 1B) (P < .001). A statistically significantly larger number of indeterminate lesions was seen on 3D FLASH (27 cases) than on SPACE (9 cases) (Fig 1C) (P = .0326), mostly related to the presence of vascular enhancement seen on 3D FLASH images.
Correlation between the number of metastatic lesions detected on axial CE 3D FLASH and SPACE (A). Differences in the number of metastatic (B) and indeterminant lesions (C) detected on the 2 sequences. Colors in A represent results obtained from different radiologists.
Figure 2 shows the distribution of the differences in the qualitative assessment results between SPACE and 3D FLASH. A positive trend was observed for all 4 metrics (image quality, lesion margin, CNR, and image artifacts), demonstrating the superiority of SPACE to 3D FLASH. Table 2 shows the results for the number of lesions, image quality, lesion margin, CNR, image artifacts, and reading time for 3D FLASH versus SPACE. Except for the quantitative CNR (n = 16), all other results include all radiologists’ evaluations (n = 92). SPACE scored significantly higher than 3D FLASH for overall image quality (P < .001), lesion margin (P < .001), CNR (P < .001), and image artifacts (P < .001).
Distribution of the differences in the qualitative assessment of image quality (A), lesion margin (B), CNR (C), and image quality (D) between axial CE 3D FLASH and SPACE.
Statistical results of detected lesions, qualitative and quantitative assessments, and reading time
A quantitative analysis of the CNR was performed in the 16 patients with solidly-enhancing lesions of >1 cm. A higher CNR was found for SPACE than for 3D FLASH images; however, this result was not statistically significant (median = 22.9 versus 15.5, respectively, P = .134). The reading time for SPACE was longer than for 3D FLASH (median = 147 seconds versus 129 seconds, respectively), and the difference was not statistically significant (P = .842).
There were high interreader lesion detection concordances for the 20 studies evaluated by 2 radiologists, resulting in Krippendorf α ordinals at 0.962 for SPACE, 0.870 for 3D FLASH, and 0.918 for the 2 sequences combined.
Evaluation of the size of enhancing lesions missed on FLASH or SPACE revealed that all except 1 lesion missed was <5 mm. The exception was a 6.5-mm ring-enhancing lesion seen only on SPACE that had the appearance of vascular enhancement on FLASH. This represented a treated metastatic lesion based on prior and subsequent MR imaging.
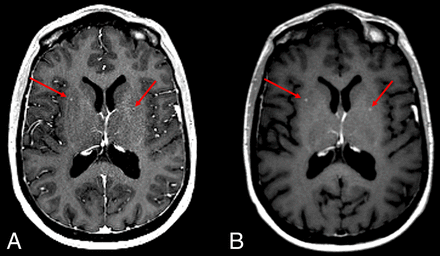
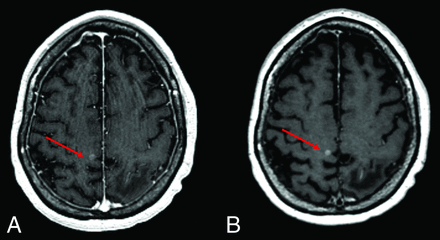
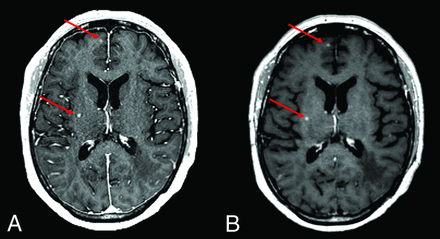
Figure 3 shows an example of CE 3D FLASH and SPACE images, with superior conspicuity of small metastases in the lentiform nucleus on the SPACE image. In the absence of vascular-enhancement suppression, these punctate foci can be falsely attributed to vascular enhancement from vessels on the 3D FLASH sequence. Figures 4 and 5 show CE 3D FLASH and SPACE images of 2 patients, with higher CNR and superior margin delineation of small metastases in the right perirolandic region and in the right basal ganglia and right frontal lobe, respectively, on SPACE images than on 3D FLASH images.
Axial CE 3D FLASH (A) and SPACE (B) images of the brain at the level of the basal ganglia demonstrate increased conspicuity of small metastases (red arrows) in the lentiform nucleus on SPACE compared with 3D FLASH.
Axial CE 3D FLASH (A) and SPACE (B) images of the brain demonstrate increased CNR and margin delineation of a small metastasis (red arrows) in the right perirolandic region on SPACE compared with 3D FLASH.
Axial CE 3D FLASH (A) and SPACE (B) images of the brain demonstrate increased CNR and margin delineation of a small metastasis (red arrows) in the right basal ganglia and right frontal lobe on SPACE compared with 3D FLASH.
DISCUSSION
CE 3D T1-weighted images are crucial to the accurate early detection of brain metastasis, with widespread implications for patient care. The ability to correctly detect the number of metastases is paramount, because it greatly affects the prognosis and choice of therapy, deciding between local therapy such as stereotactic radiosurgery or use of whole-brain radiation or systemic therapy. This study found that CE SPACE detected more lesions than 3D FLASH. This improved ability to detect CE lesions can, in part, be attributed to a higher CNR, better image quality, and fewer image artifacts on SPACE versus 3D FLASH, as shown in Table 2. There is abundant literature demonstrating that 3D FSE-based T1-weighted sequences, including SPACE, have superior lesion conspicuity and CNR compared with 3D GRE-based sequences, mostly MPRAGE.6⇓–8,13 Our findings mirror previously published data, including the results of a 2016 meta-analysis14 that found superior detection of brain metastasis with 3D FSE-based T1 sequences compared with MPRAGE in 5 published studies.6,7,12,15,16
While the MPRAGE sequence offers superior gray matter–white matter contrast as a result of the IR preparation, its detection of CE lesions may be compromised, especially in those with a low gadolinium concentration11,17 or enhancing lesions located within the bright white matter.10 Early simulation studies demonstrated higher lesion/tissue CNR with 3D FLASH than with MPRAGE, though they were both lower than a spin-echo sequence at a low gadolinium concentration, which may be improved with sequence optimization.11
There are limited studies comparing non–IR-prepared 3D GRE versus MPRAGE or SPACE for the detection of brain metastasis. Wetzel et al18 compared VIBE with MPRAGE at 1.5T and found a significantly higher SNR and CNR with VIBE in 30 patients with focal brain lesions, including 10 with suspicion of metastatic disease or CNS lymphoma. Majigsuren et al19 compared T1-Cube (GE Healthcare) and 3D fast-spoiled gradient-recalled sequences, which are equivalent to the T1 SPACE versus 3D FLASH sequences in this study but on a 3T GE scanner and found significantly higher CNRs with T1-Cube in 9 patients with brain metastasis. More recently, Danieli et al9 compared MPRAGE, SPACE, and VIBE in 54 contrast-enhancing tumors, including 16 metastases. Although both SPACE and VIBE provided significantly higher CNRs than MPRAGE, there was no significant difference between SPACE and VIBE. Note that VIBE is a similar sequence to 3D FLASH, with the main difference being the use of asymmetric k-space sampling and interpolation for acceleration.20
The present study is the first to directly demonstrate the superior performance of SPACE over a non-IR-prepared 3D GRE sequence for the detection of CE metastatic lesions in a sizable patient population (n = 72). An interesting finding in our study was that although the CNR result from the qualitative assessment significantly differed between the 2 sequences, the difference in the quantitative assessment did not reach statistical significance. This outcome may be attributed to the small sample size (n = 16) in our quantitative CNR calculation (due to the inclusion criterion of a solidly-enhancing lesion size of >1 cm) or to the small CNR difference between the 2 sequences, similar to the finding in Danieli et al.9
In this study, we found fewer indeterminate lesions using SPACE, likely due to the suppression of vascular enhancement. This metric can be used as a surrogate measure of how confident the interpreting radiologist is in the imaging findings. Small bright vessels can be difficult to differentiate from metastases, particularly in the basal ganglia or along the superficial aspect of the gyri, contributing to the decreased confidence of the interpreting radiologists. The SPACE sequence has inherent flow suppression from the intravoxel dephasing due to the uncompensated gradient moments in the echo-train and the stimulated echoes from variable flip angle radiofrequency pulses.10 Thus, enhancement can more confidently be attributed to metastatic disease. The importance of flow suppression has been further realized by improved metastasis detection using the black-blood vascular suppression (delay alternating with nutation for tailored excitation [DANTE]) version of SPACE.21 Most recently, Chkili et al22 compared the use of SPACE in combination with VIBE with SPACE alone and found that SPACE alone performed like SPACE and VIBE combined.
Improved margin delineation was seen on SPACE versus 3D FLASH (Fig 2B). CE 3D T1-weighted images are used for stereotactic radiosurgery treatment planning, in which not only lesion detection but also precise margin delineation is paramount. This improved margin delineation may result in improved targeting of radiation therapy, with downstream clinical implications. Furthermore, accurate margin delineation may improve the accuracy of lesion measurement and thus the accuracy of treatment-response assessment. A significant difference in volume measurement on SPACE versus MPRAGE was demonstrated by Danieli et al,9 with SPACE generating larger volumes. The consensus recommendation for standardized brain tumor imaging for clinical trials in brain metastases recommends using SPACE at 3T.10
We found no statistically significant differences in reading time between the 2 sequences. This finding is clinically meaningful when implementing SPACE into a busy clinical practice and is helpful for physician buy-in. The trend of slightly longer interpretation time for SPACE may be attributable to unfamiliarity with the sequence by the interpreting radiologists and possibly to more lesions being detected on SPACE, resulting in a longer time spent interpreting the study.
The use of SPACE for the detection of brain metastases has potential drawbacks related to reduced gray-white matter differentiation and the lack of vascular enhancement. The absence of enhancement of superficial venous structures may be undesirable for surgical planning.
This study has some limitations. This was a retrospective single-institution study that aimed for quality improvement. Therefore, the control condition, ie, the 3D FLASH sequence, was limited by our standard of care protocol with a different acquisition plane (axial) and voxel size (0.9 × 0.9 × 1.4 mm3) than the SPACE sequence (sagittal and 1-mm isotropic, respectively). The image review was primarily performed using axial images, which, in the case of SPACE, were reconstructed from sagittal acquisitions, while the axial 3D FLASH images represented source images. Due to the different appearances of SPACE and 3D FLASH images, it was not possible to make the qualitative assessment a blind experiment. Therefore, we implemented a 2-week separation between reviewing the 2 sequences from the same patient to at least avoid memory bias. Our study evaluated only sequence performance at 3T. A lower SNR on SPACE at 1.5T may prove challenging and will require additional studies. The detection number of metastases and indeterminate lesions was not validated with follow-up imaging to establish the accuracy of lesion detection. Without follow-up imaging, it is possible that some of the lesions included represented other etiologies such as subacute infarcts. Lesion size was not recorded, so the effect of lesion size on the performance of the sequences was not included in the primary analysis. However, the subsequent evaluation found that all except 1 lesion not detected on either of the sequences were small, <5 mm. Finally, lesion segmentation and morphometric analyses were not performed in this study, but they are important perspectives in comparing contrast-enhancing sequences in addition to the lesion detectability.
CONCLUSIONS
The CE T1-weighted SPACE sequence could detect more metastatic lesions than 3D FLASH. It was rated higher for image quality, lesion margin, and CNR and had fewer artifacts. In addition, the SPACE sequence increased reader confidence with fewer indeterminate lesions detected.
Acknowledgments
Editorial support was provided by the Research Medical Library at The University of Texas MD Anderson Cancer Center.
Footnotes
This work was conducted with support from Siemens Medical Solutions. J.P.L. received support from the National Cancer Institute and the National Center for Advancing Translational Sciences of the National Institutes of Health (P30 CA016672 and CCTS UL1TR003167).
Disclosure forms provided by the authors are available with the full text and PDF of this article at www.ajnr.org.
References
- Received August 17, 2024.
- Accepted after revision November 18, 2024.
- © 2025 by American Journal of Neuroradiology